Are you an international student? Are you interested in learning more about Genomics? Do you get overwhelmed by the amount of conflicting information you see online? If so, you need not search further because you will find the answer to that question in the article below.
To get more information on Genomics. You can also find up-to-date, related articles on Collegelearners.
Genomics is the study of all of a person’s genes (the genome), including interactions of those genes with each other and with the person’s environment.
What is DNA?
Deoxyribonucleic acid (DNA) is the chemical compound that contains the instructions needed to develop and direct the activities of nearly all living organisms. DNA molecules are made of two twisting, paired strands, often referred to as a double helix
Each DNA strand is made of four chemical units, called nucleotide bases, which comprise the genetic “alphabet.” The bases are adenine (A), thymine (T), guanine (G), and cytosine (C). Bases on opposite strands pair specifically: an A always pairs with a T; a C always pairs with a G. The order of the As, Ts, Cs and Gs determines the meaning of the information encoded in that part of the DNA molecule just as the order of letters determines the meaning of a word.
What is a genome?
An organism’s complete set of DNA is called its genome. Virtually every single cell in the body contains a complete copy of the approximately 3 billion DNA base pairs, or letters, that make up the human genome.
With its four-letter language, DNA contains the information needed to build the entire human body. A gene traditionally refers to the unit of DNA that carries the instructions for making a specific protein or set of proteins. Each of the estimated 20,000 to 25,000 genes in the human genome codes for an average of three proteins.
Located on 23 pairs of chromosomes packed into the nucleus of a human cell, genes direct the production of proteins with the assistance of enzymes and messenger molecules. Specifically, an enzyme copies the information in a gene’s DNA into a molecule called messenger ribonucleic acid (mRNA). The mRNA travels out of the nucleus and into the cell’s cytoplasm, where the mRNA is read by a tiny molecular machine called a ribosome, and the information is used to link together small molecules called amino acids in the right order to form a specific protein.
Proteins make up body structures like organs and tissue, as well as control chemical reactions and carry signals between cells. If a cell’s DNA is mutated, an abnormal protein may be produced, which can disrupt the body’s usual processes and lead to a disease such as cancer.
What is DNA sequencing?
Sequencing simply means determining the exact order of the bases in a strand of DNA. Because bases exist as pairs, and the identity of one of the bases in the pair determines the other member of the pair, researchers do not have to report both bases of the pair.
In the most common type of sequencing used today, called sequencing by synthesis, DNA polymerase (the enzyme in cells that synthesizes DNA) is used to generate a new strand of DNA from a strand of interest. In the sequencing reaction, the enzyme incorporates into the new DNA strand individual nucleotides that have been chemically tagged with a fluorescent label. As this happens, the nucleotide is excited by a light source, and a fluorescent signal is emitted and detected. The signal is different depending on which of the four nucleotides was incorporated. This method can generate ‘reads’ of 125 nucleotides in a row and billions of reads at a time.
To assemble the sequence of all the bases in a large piece of DNA such as a gene, researchers need to read the sequence of overlapping segments. This allows the longer sequence to be assembled from shorter pieces, somewhat like putting together a linear jigsaw puzzle. In this process, each base has to be read not just once, but at least several times in the overlapping segments to ensure accuracy.
Researchers can use DNA sequencing to search for genetic variations and/or mutations that may play a role in the development or progression of a disease. The disease-causing change may be as small as the substitution, deletion, or addition of a single base pair or as large as a deletion of thousands of bases.
What is the Human Genome Project?
The Human Genome Project, which was led at the National Institutes of Health (NIH) by the National Human Genome Research Institute, produced a very high-quality version of the human genome sequence that is freely available in public databases. That international project was successfully completed in April 2003, under budget and more than two years ahead of schedule.
The sequence is not that of one person, but is a composite derived from several individuals. Therefore, it is a “representative” or generic sequence. To ensure anonymity of the DNA donors, more blood samples (nearly 100) were collected from volunteers than were used, and no names were attached to the samples that were analyzed. Thus, not even the donors knew whether their samples were actually used.
The Human Genome Project was designed to generate a resource that could be used for a broad range of biomedical studies. One such use is to look for the genetic variations that increase risk of specific diseases, such as cancer, or to look for the type of genetic mutations frequently seen in cancerous cells. More research can then be done to fully understand how the genome functions and to discover the genetic basis for health and disease.
What are the implications for medical science?
Virtually every human ailment has some basis in our genes. Until recently, doctors were able to take the study of genes, or genetics, into consideration only in cases of birth defects and a limited set of other diseases. These were conditions, such as sickle cell anemia, which have very simple, predictable inheritance patterns because each is caused by a change in a single gene.
With the vast trove of data about human DNA generated by the Human Genome Project and other genomic research, scientists and clinicians have more powerful tools to study the role that multiple genetic factors acting together and with the environment play in much more complex diseases. These diseases, such as cancer, diabetes, and cardiovascular disease constitute the majority of health problems in the United States. Genome-based research is already enabling medical researchers to develop improved diagnostics, more effective therapeutic strategies, evidence-based approaches for demonstrating clinical efficacy, and better decision-making tools for patients and providers. Ultimately, it appears inevitable that treatments will be tailored to a patient’s particular genomic makeup. Thus, the role of genetics in health care is starting to change profoundly and the first examples of the era of genomic medicine are upon us.
It is important to realize, however, that it often takes considerable time, effort, and funding to move discoveries from the scientific laboratory into the medical clinic. Most new drugs based on genome-based research are estimated to be at least 10 to 15 years away, though recent genome-driven efforts in lipid-lowering therapy have considerably shortened that interval. According to biotechnology experts, it usually takes more than a decade for a company to conduct the kinds of clinical studies needed to receive approval from the Food and Drug Administration.
Screening and diagnostic tests, however, are here. Rapid progress is also being made in the emerging field of pharmacogenomics, which involves using information about a patient’s genetic make-up to better tailor drug therapy to their individual needs.
Clearly, genetics remains just one of several factors that contribute to people’s risk of developing most common diseases. Diet, lifestyle, and environmental exposures also come into play for many conditions, including many types of cancer. Still, a deeper understanding of genetics will shed light on more than just hereditary risks by revealing the basic components of cells and, ultimately, explaining how all the various elements work together to affect the human body in both health and disease.
Genomics: a revolution in health care?
The Holy Grail in health care has long been personalized medicine, or what is now called precision medicine,” says Kemal Malik, member of the Bayer board of management responsible for innovation. “But getting to the level of precision we wanted wasn’t possible until now. What’s changed is our ability to sequence the human genome.” In April 2003, the Human Genome Project announced that it had sequenced around 20,000 genes of those that make up the blueprint of our bodies. For 15 years, this medical breakthrough has been informing and transforming health care. Genomics, the study of genes, is making it possible to predict, diagnose, and treat diseases more precisely and personally than ever.
A complete human genome contains three billion base pairs of DNA, uniquely arranged to give us our fundamental anatomy and individual characteristics such as height and hair color. DNA forms genes and understanding their function gives crucial insights into how our bodies work and what happens when we get sick. This was the reasoning behind the 13 years and $2.7 billion spent on the Human Genome Project. The world has quickly built on its achievements and now we can map a human genome in just a few hours and for less than a thousand dollars. Fast, large-scale, low-cost DNA sequencing has propelled genomics into mainstream medicine, driving a revolutionary shift toward precision medicine.
Early diagnosis of a disease can significantly increase the chances of successful treatment, and genomics can detect a disease long before symptoms present themselves. Many diseases, including cancers, are caused by alterations in our genes. Genomics can identify these alterations and search for them using an ever-growing number of genetic tests, many available online. If your results suggest susceptibility to a condition, you may be able to take preemptive action to delay or even stop the disease developing. “Health care will move more toward prevention rather than cure,” Malik says, and genomics will likely prove an important enabler in understanding the particular healthcare steps an individual should or should not take.
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism’s complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of individual genes and their roles in inheritance, genomics aims at the collective characterization and quantification of all of an organism’s genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in genomics have triggered a revolution in discovery-based research and systems biology to facilitate understanding of even the most complex biological systems such as the brain.
The field also includes studies of intragenomic (within the genome) phenomena such as epistasis (effect of one gene on another), pleiotropy (one gene affecting more than one trait), heterosis (hybrid vigour), and other interactions between loci and alleles within the genome
History
Etymology
From the Greek ΓΕΝ gen, “gene” (gamma, epsilon, nu, epsilon) meaning “become, create, creation, birth”, and subsequent variants: genealogy, genesis, genetics, genic, genomere, genotype, genus etc. While the word genome (from the German Genom, attributed to Hans Winkler) was in use in English as early as 1926, the term genomics was coined by Tom Roderick, a geneticist at the Jackson Laboratory (Bar Harbor, Maine), over beer at a meeting held in Maryland on the mapping of the human genome in 1986.
Early sequencing efforts
Following Rosalind Franklin’s confirmation of the helical structure of DNA, James D. Watson and Francis Crick’s publication of the structure of DNA in 1953 and Fred Sanger’s publication of the Amino acid sequence of insulin in 1955, nucleic acid sequencing became a major target of early molecular biologists. In 1964, Robert W. Holley and colleagues published the first nucleic acid sequence ever determined, the ribonucleotide sequence of alanine transfer RNA. Extending this work, Marshall Nirenberg and Philip Leder revealed the triplet nature of the genetic code and were able to determine the sequences of 54 out of 64 codons in their experiments. In 1972, Walter Fiers and his team at the Laboratory of Molecular Biology of the University of Ghent (Ghent, Belgium) were the first to determine the sequence of a gene: the gene for Bacteriophage MS2 coat protein. Fiers’ group expanded on their MS2 coat protein work, determining the complete nucleotide-sequence of bacteriophage MS2-RNA (whose genome encodes just four genes in 3569 base pairs [bp]) and Simian virus 40 in 1976 and 1978, respectively.
DNA-sequencing technology developed
Walter GilbertFrederick Sanger and Walter Gilbert shared half of the 1980 Nobel Prize in Chemistry for Independently developing methods for the sequencing of DNA.
In addition to his seminal work on the amino acid sequence of insulin, Frederick Sanger and his colleagues played a key role in the development of DNA sequencing techniques that enabled the establishment of comprehensive genome sequencing projects. In 1975, he and Alan Coulson published a sequencing procedure using DNA polymerase with radiolabelled nucleotides that he called the Plus and Minus technique. This involved two closely related methods that generated short oligonucleotides with defined 3′ termini. These could be fractionated by electrophoresis on a polyacrylamide gel (called polyacrylamide gel electrophoresis) and visualised using autoradiography. The procedure could sequence up to 80 nucleotides in one go and was a big improvement, but was still very laborious. Nevertheless, in 1977 his group was able to sequence most of the 5,386 nucleotides of the single-stranded bacteriophage φX174, completing the first fully sequenced DNA-based genome. The refinement of the Plus and Minus method resulted in the chain-termination, or Sanger method (see below), which formed the basis of the techniques of DNA sequencing, genome mapping, data storage, and bioinformatic analysis most widely used in the following quarter-century of research. In the same year Walter Gilbert and Allan Maxam of Harvard University independently developed the Maxam-Gilbert method (also known as the chemical method) of DNA sequencing, involving the preferential cleavage of DNA at known bases, a less efficient method. For their groundbreaking work in the sequencing of nucleic acids, Gilbert and Sanger shared half the 1980 Nobel Prize in chemistry with Paul Berg (recombinant DNA).
Complete genomes
The advent of these technologies resulted in a rapid intensification in the scope and speed of completion of genome sequencing projects. The first complete genome sequence of a eukaryotic organelle, the human mitochondrion (16,568 bp, about 16.6 kb [kilobase]), was reported in 1981, and the first chloroplast genomes followed in 1986. In 1992, the first eukaryotic chromosome, chromosome III of brewer’s yeast Saccharomyces cerevisiae (315 kb) was sequenced. The first free-living organism to be sequenced was that of Haemophilus influenzae (1.8 Mb [megabase]) in 1995. The following year a consortium of researchers from laboratories across North America, Europe, and Japan announced the completion of the first complete genome sequence of a eukaryote, S. cerevisiae (12.1 Mb), and since then genomes have continued being sequenced at an exponentially growing pace. As of October 2011, the complete sequences are available for: 2,719 viruses, 1,115 archaea and bacteria, and 36 eukaryotes, of which about half are fungi.The number of genome projects has increased as technological improvements continue to lower the cost of sequencing. (A) Exponential growth of genome sequence databases since 1995. (B) The cost in US Dollars (USD) to sequence one million bases. (C) The cost in USD to sequence a 3,000 Mb (human-sized) genome on a log-transformed scale.
Most of the microorganisms whose genomes have been completely sequenced are problematic pathogens, such as Haemophilus influenzae, which has resulted in a pronounced bias in their phylogenetic distribution compared to the breadth of microbial diversity. Of the other sequenced species, most were chosen because they were well-studied model organisms or promised to become good models. Yeast (Saccharomyces cerevisiae) has long been an important model organism for the eukaryotic cell, while the fruit fly Drosophila melanogaster has been a very important tool (notably in early pre-molecular genetics). The worm Caenorhabditis elegans is an often used simple model for multicellular organisms. The zebrafish Brachydanio rerio is used for many developmental studies on the molecular level, and the plant Arabidopsis thaliana is a model organism for flowering plants. The Japanese pufferfish (Takifugu rubripes) and the spotted green pufferfish (Tetraodon nigroviridis) are interesting because of their small and compact genomes, which contain very little noncoding DNA compared to most species. The mammals dog (Canis familiaris) brown rat (Rattus norvegicus), mouse (Mus musculus), and chimpanzee (Pan troglodytes) are all important model animals in medical research.
A rough draft of the human genome was completed by the Human Genome Project in early 2001, creating much fanfare. This project, completed in 2003, sequenced the entire genome for one specific person, and by 2007 this sequence was declared “finished” (less than one error in 20,000 bases and all chromosomes assembled). In the years since then, the genomes of many other individuals have been sequenced, partly under the auspices of the 1000 Genomes Project, which announced the sequencing of 1,092 genomes in October 2012. Completion of this project was made possible by the development of dramatically more efficient sequencing technologies and required the commitment of significant bioinformatics resources from a large international collaboration. The continued analysis of human genomic data has profound political and social repercussions for human societies.
The “omics” revolution
General schema showing the relationships of the genome, transcriptome, proteome, and metabolome (lipidome).Main articles: Omics and Human proteome project
The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics. The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively. The suffix -ome as used in molecular biology refers to a totality of some sort; similarly omics has come to refer generally to the study of large, comprehensive biological data sets. While the growth in the use of the term has led some scientists (Jonathan Eisen, among others) to claim that it has been oversold, it reflects the change in orientation towards the quantitative analysis of complete or near-complete assortment of all the constituents of a system. In the study of symbioses, for example, researchers which were once limited to the study of a single gene product can now simultaneously compare the total complement of several types of biological molecules.
Genome analysis
After an organism has been selected, genome projects involve three components: the sequencing of DNA, the assembly of that sequence to create a representation of the original chromosome, and the annotation and analysis of that representation.Overview of a genome project. First, the genome must be selected, which involves several factors including cost and relevance. Second, the sequence is generated and assembled at a given sequencing center (such as BGI or DOE JGI). Third, the genome sequence is annotated at several levels: DNA, protein, gene pathways, or comparatively.
Sequencing
Historically, sequencing was done in sequencing centers, centralized facilities (ranging from large independent institutions such as Joint Genome Institute which sequence dozens of terabases a year, to local molecular biology core facilities) which contain research laboratories with the costly instrumentation and technical support necessary. As sequencing technology continues to improve, however, a new generation of effective fast turnaround benchtop sequencers has come within reach of the average academic laboratory. On the whole, genome sequencing approaches fall into two broad categories, shotgun and high-throughput (or next-generation) sequencing.
Shotgun sequencing
An ABI PRISM 3100 Genetic Analyzer. Such capillary sequencers automated early large-scale genome sequencing efforts.Main article: Shotgun sequencing
Shotgun sequencing is a sequencing method designed for analysis of DNA sequences longer than 1000 base pairs, up to and including entire chromosomes. It is named by analogy with the rapidly expanding, quasi-random firing pattern of a shotgun. Since gel electrophoresis sequencing can only be used for fairly short sequences (100 to 1000 base pairs), longer DNA sequences must be broken into random small segments which are then sequenced to obtain reads. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence. Shotgun sequencing is a random sampling process, requiring over-sampling to ensure a given nucleotide is represented in the reconstructed sequence; the average number of reads by which a genome is over-sampled is referred to as coverage.
For much of its history, the technology underlying shotgun sequencing was the classical chain-termination method or ‘Sanger method’, which is based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. Recently, shotgun sequencing has been supplanted by high-throughput sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use, primarily for smaller-scale projects and for obtaining especially long contiguous DNA sequence reads (>500 nucleotides). Chain-termination methods require a single-stranded DNA template, a DNA primer, a DNA polymerase, normal deoxynucleosidetriphosphates (dNTPs), and modified nucleotides (dideoxyNTPs) that terminate DNA strand elongation. These chain-terminating nucleotides lack a 3′-OH group required for the formation of a phosphodiester bond between two nucleotides, causing DNA polymerase to cease extension of DNA when a ddNTP is incorporated. The ddNTPs may be radioactively or fluorescently labelled for detection in DNA sequencers. Typically, these machines can sequence up to 96 DNA samples in a single batch (run) in up to 48 runs a day.
High-throughput sequencing
The high demand for low-cost sequencing has driven the development of high-throughput sequencing technologies that parallelize the sequencing process, producing thousands or millions of sequences at once. High-throughput sequencing is intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods. In ultra-high-throughput sequencing, as many as 500,000 sequencing-by-synthesis operations may be run in parallel.Illumina Genome Analyzer II System. Illumina technologies have set the standard for high-throughput massively parallel sequencing.[
The Illumina dye sequencing method is based on reversible dye-terminators and was developed in 1996 at the Geneva Biomedical Research Institute, by Pascal Mayer [fr] and Laurent Farinelli. In this method, DNA molecules and primers are first attached on a slide and amplified with polymerase so that local clonal colonies, initially coined “DNA colonies”, are formed. To determine the sequence, four types of reversible terminator bases (RT-bases) are added and non-incorporated nucleotides are washed away. Unlike pyrosequencing, the DNA chains are extended one nucleotide at a time and image acquisition can be performed at a delayed moment, allowing for very large arrays of DNA colonies to be captured by sequential images taken from a single camera. Decoupling the enzymatic reaction and the image capture allows for optimal throughput and theoretically unlimited sequencing capacity; with an optimal configuration, the ultimate throughput of the instrument depends only on the A/D conversion rate of the camera. The camera takes images of the fluorescently labeled nucleotides, then the dye along with the terminal 3′ blocker is chemically removed from the DNA, allowing the next cycle.
An alternative approach, ion semiconductor sequencing, is based on standard DNA replication chemistry. This technology measures the release of a hydrogen ion each time a base is incorporated. A microwell containing template DNA is flooded with a single nucleotide, if the nucleotide is complementary to the template strand it will be incorporated and a hydrogen ion will be released. This release triggers an ISFET ion sensor. If a homopolymer is present in the template sequence multiple nucleotides will be incorporated in a single flood cycle, and the detected electrical signal will be proportionally higher.
Assembly
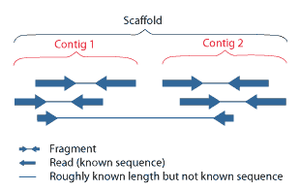
Overlapping reads form contigs; contigs and gaps of known length form scaffolds.
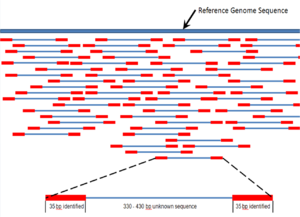
Paired end reads of next generation sequencing data mapped to a reference genome.Multiple, fragmented sequence reads must be assembled together on the basis of their overlapping areas.
Sequence assembly refers to aligning and merging fragments of a much longer DNA sequence in order to reconstruct the original sequence. This is needed as current DNA sequencing technology cannot read whole genomes as a continuous sequence, but rather reads small pieces of between 20 and 1000 bases, depending on the technology used. Third generation sequencing technologies such as PacBio or Oxford Nanopore routinely generate sequencing reads >10 kb in length; however, they have a high error rate at approximately 15 percent. Typically the short fragments, called reads, result from shotgun sequencing genomic DNA, or gene transcripts (ESTs).
Assembly approaches
Assembly can be broadly categorized into two approaches: de novo assembly, for genomes which are not similar to any sequenced in the past, and comparative assembly, which uses the existing sequence of a closely related organism as a reference during assembly. Relative to comparative assembly, de novo assembly is computationally difficult (NP-hard), making it less favourable for short-read NGS technologies. Within the de novo assembly paradigm there are two primary strategies for assembly, Eulerian path strategies, and overlap-layout-consensus (OLC) strategies. OLC strategies ultimately try to create a Hamiltonian path through an overlap graph which is an NP-hard problem. Eulerian path strategies are computationally more tractable because they try to find a Eulerian path through a deBruijn graph.
Finishing
Finished genomes are defined as having a single contiguous sequence with no ambiguities representing each replicon.
Annotation
The DNA sequence assembly alone is of little value without additional analysis. Genome annotation is the process of attaching biological information to sequences, and consists of three main steps:
- identifying portions of the genome that do not code for proteins
- identifying elements on the genome, a process called gene prediction, and
- attaching biological information to these elements.
Automatic annotation tools try to perform these steps in silico, as opposed to manual annotation (a.k.a. curation) which involves human expertise and potential experimental verification. Ideally, these approaches co-exist and complement each other in the same annotation pipeline (also see below).
Traditionally, the basic level of annotation is using BLAST for finding similarities, and then annotating genomes based on homologues. More recently, additional information is added to the annotation platform. The additional information allows manual annotators to deconvolute discrepancies between genes that are given the same annotation. Some databases use genome context information, similarity scores, experimental data, and integrations of other resources to provide genome annotations through their Subsystems approach. Other databases (e.g. Ensembl) rely on both curated data sources as well as a range of software tools in their automated genome annotation pipeline. Structural annotation consists of the identification of genomic elements, primarily ORFs and their localisation, or gene structure. Functional annotation consists of attaching biological information to genomic elements.
When symptoms do develop, genomics can be instrumental in diagnosing the problem. The Human Genome Project has fueled the discovery of nearly 2,000 disease genes, and these are proving highly effective at providing fast and accurate analysis. They have been especially valuable for identifying rare genetic diseases that had previously taken years to diagnose, ending the uncertainty and suffering of “diagnostic odysseys.” Ongoing research is committed to building databases of genetic biomarkers, especially for cancers. “To date, genomics has had the most impact on cancer,” Malik explains, “because we can get tissue, sequence it, and identify the alterations.” In the United States, the Cancer Genome Atlas has mapped the key genomic changes in more than 30 types of cancer. Such databases could deliver a definitive diagnosis in seconds, and even recommend targeted treatments based on the DNA of both the patient and the disease. Indeed, genetic sequencing of cancer tumors is helping not only to identify particular cancers but also to understand what causes them and what could kill them.
When it comes to treatment, genomics is driving another important element of precision medicine—pharmacogenomics. “We’ve long known that the same medication in the same dose affects people differently,” Malik says. Now we know why: Our genes influence the production of crucial enzymes in the liver that metabolize medicines. If a genetic variation stops the enzymes from working properly, the drug can build up in the body with serious side effects. Other drugs only work when broken down in the liver, so if the enzymes don’t work, neither does the drug. Such gene variations, known as polymorphisms, are common, but genomics means we can test for them and compensate for them. Gene variations mean that around 30 percent of people cannot fully convert a commonly used anti-clotting drug, but gene testing means alternative drugs can be taken to the same effect.
There are more than 250 drugs labeled with pharmacogenomic information, enabling them to be prescribed based on a patient’s genetics. As the number grows, and as DNA sequencing becomes standard, it’s likely that medicines will be routinely prescribed based on our genes—minimizing harmful side effects and making treatments faster and more effective. “Genomics is even changing the way we develop drugs,” Malik says. “It’s much more sophisticated because we can find a specific alteration that is causing a disease and try to target a medicine specifically to that alteration.” In 2014 eight out of the 41 new drugs approved by the FDA were targeted therapies. “Traditionally, health care’s approach has been a bit general,” Malik says. “Now we can look at a disease on a personal level.”
Nowhere is this more palpable than in the emerging field of gene editing, a technique that sees scientists change the DNA of living organisms. “Seldom does medicine really get to cure people,” Malik says. “But if the underlying problem is a particular gene alteration and we can snip that alteration out, then we’ve shifted from treating to curing.” A key technology behind gene editing is CRISPR-Cas9. Based on a defense mechanism found in bacteria, the Cas9 enzyme, described by Malik as “molecular scissors,” is delivered to a precise segment of malfunctioning DNA, cuts it out, and replaces it with good DNA created in the lab. Already in clinical trials, gene editing is becoming a fast, effective, and increasingly affordable treatment. “This is the ultimate precision medicine,” he says. “It has huge potential.”
In the 15 years since sequencing the first human genome, medicine has been quick to put its learnings into action. Genomics is providing us with a human instruction manual that is showing us how to fix ourselves. “In the future we’ll see every cancer patient sequenced, and we’ll develop specific drugs to target their particular genetic alteration,” Malik suggests. With DNA sequencing heading below the $100 mark, by 2025 as many as two billion genomes could have been sequenced. “We’re moving very, very rapidly,” he says. Genomics is putting the patient rather than the disease at the heart of health care, with a shift from treatments to cures. “It’s revolutionizing people’s ideas not just of health care,” Malik says, “but of illness itself.”
The next chapter for African genomics
In the affluent, beach-side neighbourhoods of Lagos, finance and technology entrepreneurs mingle with investors at art openings and chic restaurants. Now biotech is entering the scene. Thirty-four-year-old Abasi Ene-Obong has been traversing the globe for the past six months, trying to draw investors and collaborators into a venture called 54Gene. Named to reflect the 54 countries in Africa, the genetics company aims to build the continent’s largest biobank, with backing from Silicon Valley venture firms such as Y Combinator and Fifty Years. The first step in that effort is a study, launched earlier this month, to sequence and analyse the genomes of 100,000 Nigerians.
At a trendy African fusion restaurant, Ene-Obong is explaining how the company can bring precision medicine to Nigeria, and generate a profit at the same time. He talks about some new investors and partners that he’s not able to name publicly, then pulls out his phone to show pictures of a property he just purchased to expand the company’s lab space.
“My big-picture vision is that we can be a reason that new drugs are discovered,” Ene-Obong says. “I don’t want science for the sake of science, I want to do science to solve problems.”Ancient African genomes offer glimpse into early human history
It’s too soon to say whether he will succeed. But his ambitions would have been unthinkable a decade ago, when most universities and hospitals in Nigeria lacked even the most basic tools for modern genetics research. Ene-Obong, the chief executive of 54Gene, is riding a wave of interest and investment in African genomics that is coursing through Nigeria. In a rural town in the western part of the country, a microbiologist is constructing a US$3.9-million genomics centre. And in the capital city of Abuja, researchers are revamping the National Reference Laboratory to analyse DNA from 200,000 blood samples stored in their new biobank. Studying everything from diabetes to cholera, these endeavours are designed to build the country’s capabilities so that genetics results from Africa — the publications, patents, jobs and any resulting therapies — flow back to the continent.
The rest of the world is interested, too. Africa contains much more genetic diversity than any other continent because humans originated there. This diversity can provide insights into human evolution and common diseases. Yet fewer than 2% of the genomes that have been analysed come from Africans. A dearth of molecular-biology research on the continent also means that people of African descent might not benefit from drugs tailored to unique genetic variations. Infectious-disease surveillance also falls short, meaning that dangerous pathogens could evade detection until an outbreak is too big to contain easily.
But Nigeria’s genetics revolution could just as soon sputter as soar. Although the country is Africa’s largest economy, its research budget languishes at 0.2% of gross domestic product (GDP). Biologists therefore need to rely on private investment or on funding from outside Africa. This threatens continuity: one of the largest US grants to Nigerian geneticists, through a project known as H3Africa, is set to expire in two years. There are other challenges. Human research in Africa requires copious communication and unique ethical consideration given the vast economic disparities and history of exploitation on the continent. And a lack of reliable electricity in Nigeria hobbles research that relies on sub-zero freezers, sensitive equipment and computing power.

Yet with a hustle that Nigerians are famous for, scientists are pushing ahead. Ene-Obong hopes to pursue research through partnerships with pharmaceutical companies, and other geneticists are competing for international grants and collaborations, or looking to charge for biotech services that are usually provided by labs outside Africa. Last November, Nnaemeka Ndodo, chief molecular bioengineer at the National Reference Laboratory, launched the Nigerian Society of Human Genetics in the hope of bringing scientists together. “When I look at the horizon it looks great — but in Nigeria you can never be sure,” he says.
Building the foundation
Around 15 years ago, Nigerian geneticist Charles Rotimi was feeling dismayed. He was enjoying academic success, but would have preferred to do so in his home country. He had left Africa to do cutting-edge research, and he was not alone.
Many Nigerian academics move abroad. According to the Migration Policy Institute in Washington DC, 29% of Nigerians aged 25 or older in the United States hold a master’s or a doctoral degree, compared with 11% of the general US population.
After Rotimi joined the US National Institutes of Health (NIH) in Bethesda, Maryland, in 2008, he hatched a plan with director Francis Collins to drive genetics research in Africa. Rotimi wasn’t interested in one-off grants, but rather in building a foundation on which science could thrive. “The major thing to me was to create jobs so that people could do the work locally,” he says. In 2010, the NIH and Wellcome, a biomedical charity in London, announced the H3Africa, or Human Heredity and Health in Africa, project. It’s become a $150-million, 10-year initiative that supports institutes in 12 African countries. The proof of its success will be not in the number of papers published, but rather in the number of African investigators able to charge ahead after the grant ends in 2022.Genomics is failing on diversity
For that to happen, H3Africa researchers realized they needed to revise research regulations and procedures for gaining the public’s trust. So rather than just collecting blood and leaving — the approach disparagingly referred to as helicopter research — many investigators on the team have devoted time to adapting studies for the African context.
For example, when Mayowa Owolabi, a neurologist at the University of Ibadan, Nigeria, was recruiting healthy controls for his H3Africa study on the genetics of stroke, his team discovered that many people had alarmingly high blood pressure and didn’t know it. Nigeria has one of the world’s highest stroke rates, and Owolabi realized that communities needed medical information and basic care more urgently than genetics. So he extended his study to include education on exercise, smoking and diet. And, on finding that many people had never heard of genetics, the team attempted to explain the concept.
This is a continuing process. One morning last November — seven years into the project — a community leader in Ibadan visited Owolabi’s private clinic. He said tensions had mounted because people who had participated in the study wanted to know the results of their genetic tests. Owolabi replied that they were still searching for genetic markers that would reveal a person’s risk of stroke, and that it might be many years until any were found. “But it’s a heart-warming question,” he says, “because if the people demand a test, it means the study is the right thing to do.”

Discovering the genetic underpinnings of stroke is also complicated by the fact that it, like many non-communicable disorders, is caused by a blend of biological and environmental factors. Owolabi flips through a blue booklet of questions answered by 9,000 participants so far. It asks about everything from family medical history to level of education. Insights are buried in the answers, even without DNA data: the team found, for instance, that young Nigerians and Ghanaians who eat green leafy vegetables every day have fewer strokes1. And that’s just the beginning. “You see the amount of data we have accrued,” he says. “I don’t think we have used even 3% of it, so we need to get more funding to keep the work going.”
Owolabi’s team is now applying for new grants from the NIH, Wellcome and other international donors to sustain the work after the H3Africa grant ends. And to make themselves more appealing to collaborators and donors, they’re increasing the amount of work they can do in Ibadan. Until last year, most of the genetic analyses were conducted at the University of Alabama in Tuscaloosa. But last June, the University of Ibadan installed a computer cluster to serve the project, and three young bioinformaticians are now crunching the data. “The big-data business is happening now,” says Adigun Taiwo Olufisayo, a doctoral student concentrating on bioinformatics. But he also admits that funds are tight.
Last year, other graduate students on the team began to extract DNA from samples so that they can scour it for genetic variants linked to strokes. In a room the size of a cupboard, a technician labels tubes beside a freezer. Coker Motunrayo, a doctoral student studying memory loss after strokes, sits on the counter-top because there’s not enough space for a chair. She insists that the H3Africa project is a success, even though their genetics work has just started. “Compare this to where we were five years ago, and you’d be stunned,” she says.
On the cusp
Perhaps the most advanced genomics facility in West Africa right now is located in Ede in southwestern Nigeria. At Redeemer’s University, a private institution founded by a Nigerian megachurch, microbiologist Christian Happi is building an empire. Construction teams are busy creating a $3.9-million home for the African Centre of Excellence for Genomics of Infectious Diseases.
Happi strides across a veranda, and into a series of rooms that will become a high-level biosafety laboratory suitable for working on Ebola and other dangerous pathogens. Another small room nearby will house a NovaSeq 6000 machine made by Illumina in San Diego, California, a multimillion-dollar piece of equipment that can sequence an entire human genome in less than 12 hours. It’s the first of that model on the continent, says Happi, and it positions his centre, and Africa, “to become a player in the field of precision medicine”. Then he announces that Herman Miller furniture is on the way. If it’s good enough for his collaborators at the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, he adds, it is good enough for his team.

Happi plans to move his lab into the facility in a few months. But the team is already doing advanced work on emerging outbreaks. At a small desk, one of Happi’s graduate students, Judith Oguzie, stares at an interactive pie chart on her laptop. The chart displays all of the genetic sequences recovered from a blood sample shipped to the lab from a hospital as part of a countrywide effort to learn which microbes are infecting people with fevers. Typically, doctors test the patients for the disease they think is most likely, such as malaria, but this means other infections can be missed. For example, the sequences Oguzie is looking at belong to the Plasmodium parasites that cause malaria, the virus that causes the deadly Lassa fever, and human papillomavirus.
Oguzie says that a few years ago, she was processing samples from a hospital in which people were dying because their fevers had confounded diagnosis. With the help of next-generation sequencing, she found that they were infected with the virus that causes yellow fever. She showed Happi the results, and he reported the news to the Nigeria Centre for Disease Control (NCDC), which rapidly launched a vaccination campaign.
This was exactly what Oguzie had wanted out of science. “I’m happy when I solve problems that have to do with life,” she says. She worked hard throughout university in Borno, even after the terrorist organization Boko Haram started attacking the northern state. She heard bomb blasts during lectures and knew people who were shot.
The effect of bias in genomic studies
The field of genomics appears to be shrouded by bias at different levels. Racial bias exists in genomic databases, making it more difficult and expensive to diagnose genetic diseases and employ strategies for treatment in African–American patients.
Historical bias also exists, with researchers less likely to study genes that are potentially implicated in diseases if those genes are less well-researched.
Historical bias
Genes that have been shown to have potentially significant implications in human disease are being ignored due to historical bias. Previous studies have reported that research is focused around only 2000 coding genes, out of a possible 20,000 genes in the human genome. Now a study, published recently in PLOS Biology, addresses the reasoning behind why biomedical researchers continually study the same 10% of human genes.
The team discovered that historical bias played a huge role in this, with old policies, funding and career paths being the main driving forces behind this.
“We discovered that current research on human genes does not reflect the medical importance of the gene,” commented Thomas Stoeger (Northwestern University; IL, USA). “Many genes with a very strong relevance to human disease are still not studied. Instead, social forces and funding mechanisms reinforce a focus of present-day science on past research topics.”
The researchers were able to analyze approximately 15,000 genes; applying a systems approach to the data to uncover underlying patterns. The result is not only an explanation of why certain genes are not studied, but also an explanation of the extent to which a gene has been studied.
They discovered that post-docs and PhD students that focus on less well-categorized genes have a 50% less chance of becoming an independent researcher. As well as this, policies that were implemented in order to further innovative research have actually just resulted in more research on well-categorized genes – the small group of genes that were the focus of much research prior to the Human Genome Project.
Since the completion of the Human Genome Project in 2003, there have been many novel technologies developed to study genes. However, studies on less than 10% of genes comprise more than 90% of research papers and approximately 30% of genes have not been studied at all. One of the key goals of the Human Genome Project was to expand study of the human genome beyond the small group of genes scientists at the time were continuously studying. Given these revelations, it seems as though the project has somewhat failed.
“Everything was supposed to change with the Human Genome Project, but everything stayed the same,” said Luis Amaral (Northwestern University). “Scientists keep going to the same place, striding the exact same genes. Should we be focusing all of our attention on this small group of genes?”
“The bias to study the exact same human genes is very high,” continued Amaral. “The entire system is fighting the very purpose of the agencies and scientific knowledge which is to broaden the set of things we study and understand. We need to make a concerted effort to incentivize the study of other genes important to human health.”
The Northwestern team will now utilize their research to build a public database identifying genes that have been less well studied but could be importantly implicated in human disease.
Racial bias
It has previously been shown that two of the top genetic databases contain considerably more genetic data on those of European descent than those of African descent. When the researchers compared their own dataset of 642 whole-genome sequences from those of African ancestry to current genomic databases, they discovered that there was a clear preference for European genetic variants.
“The ability to accurately report whether a genetic variant is responsible for a given disease or phenotypic trait depends in part on the confidence in labelling a variant as pathogenic,” explained the authors of the paper, published in Nature Communications. “Such determination can often be more difficult in persons of predominantly non-European ancestry, as there is less known about the pathogenicity of variants that are absent from or less frequent in European populations.”
In the study, the researchers distinguished pathogenic annotated variants, which have been identified as disease-causing on online databases, from non-annotated variants (NAVs), which are not identified as disease-causing.
“While we cannot be sure which of these variants are truly disease-causing (actual ‘needles’ rather than haystack members) without additional functional or association-based evidence, we believe that discrepancies between true pathogenicity and annotated pathogenicity are a major source of the biases we report,” the authors commented. “A likely contributor to this incongruity is that databases are missing population-specific pathogenicity information, and with regard to the results we report here, African-specific pathogenicity data.”
The researchers discovered that NAVs have the highest degree of positive correlation with those of African ancestry. Therefore, genetic variants that are disease-causing for this population (the needles) are likely to be in the NAV category (the haystack), and as such, will be harder to find. This is due to the sheer volume of NAVs, which will be even greater for those of African descent, who have a larger degree of genetic variation.
The databases must now be expanded to include a wider range of ancestries. This will dramatically change clinical genetics and diagnoses for African–American patients. Currently, the task of analyzing these patients’ genomes is a more difficult and expensive one, compared with those of European descent.
Although these biases may seem like tough fixes to make, there is hope that awareness will set in action the steps that need to be implemented to overcome them and move research forward.